Last week's molecule was tetrahydrofolate, an essential cofactor in several reactions; notably the synthesis of thymidine. The winner was Jacob Troth [Monday's Molecule #195]. He should contact me by email.
This week's molecule is a very important molecule. I'm showing you two different conformations of the same molecule. You need to identify this molecule using its full and complete common name. I'm going to be strict about this, if you give me an ambiguous name you will not win.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
Last week's molecule was allantoin, the breakdown product of uric acid that's found in the urine of mammals. Other animals can degrade allantoin to more simple compounds but mammals have lost the genes for this pathway. The winners were Paul Clapham and Jacob Troth [Monday's Molecule #194]. They should contact me by email.
This week's molecule is a ubiquitous and essential molecule in all species. Give the common name but be sure you don't confuse it with other, very similar, molecules.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
One of the remarkable things about kooks is that they are incredibly resistant to learning from their mistakes. James Shapiro gives us a fine example in one of his latest articles on The Huffington Post where he tries to convince us that the old definition of "gene" has outlived its usefulness. According to Shapiro, "DNA and molecular genetics have brought us to a fundamentally new conceptual understanding of genomes, how they are organized and how they function."
Really? While we all can agree that there's no definition of "gene" that doesn't have exceptions, we can surely agree that some definitions work pretty well. I've argued that defining a gene as, "a DNA sequence that is transcribed to produce a functional product" works well in most cases [What Is a Gene?].
Let's see how James Shapiro handles this problem.1 He says,
The identification of DNA as the key molecule of heredity and Crick's Central Dogma of Molecule Biology [Crick 1970] initially seemed to confirm Beadle and Tatum's "one gene -- one enzyme" hypothesis.
I've already explained that Shapiro doesn't understand the Central Dogma of Molecular Biology even though he quotes the Francis Crick papers that explain it correctly [Revisiting the Central Dogma in the 21st Century]. I also made this point in my review of his book: Evolution: A View from the 21st Century.
Read more »
Last week's molecule was N-(2-aminoethyl)glycine, a molecule that has been used to build artificial DNA molecules. The winner was Michael Rasmussen [Monday's Molecule #193].
This week's molecule is a real pisser. You'll have to give the complete, unambiguous, formal name AND explain why we don't make it.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
Last week's molecule was capsaicin, the molecule responsible for the "hot" sensation of chili peppers. There were two winners: Seth Kasowitz and Bill Gunn [Monday's Molecule #192].
This week's molecule is featured in an article that I will (hopefully) blog about in the next few days. There's a common name of sorts but you will need to supply the correct IUPAC name to win the free lunch.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
Last week's molecule was photosystem I (PSI). Mikkel Rasmussen was the only one to get it right [Monday's Molecule #191].
I thought of this week's molecule while I was in Los Angeles last week. We got to sample some excellent examples of cuisine that's hard to find in Toronto. You need to supply the common name AND the formal IUPAC name.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
Last week's molecule was L-dopa. The winner was Raul A. Félix de Sousa (again, but this time only by four minutes!) [Monday's Molecule #190].
This week's molecule is much more complicated and it's also much more important. You need to identify this complex making sure you distinguish it from other similar complexes. You don't need to name the exact species but you should have some idea of which organisms have this complex and which ones don't. There's not enough room in the comments for the complete IUPAC name!
You'll get special bonus points (and the expensive lunch in the dining room instead of the pub) for explaining how an irreducibly complex structure like this could have evolved.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
Last week's molecules were cis Δ9-octadecanic acid (oleic acid) and trans Δ9-octadecanic acid (elaidic acid). Last week's winner was John Runnels. He should email me.
Name this week's molecule. Be sure to give an unambiguous name—it can be the common name or the IUPAC name. Why is this molecule important in some species?
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
There's been a lot of talk recently about the discussion on reddit concerning the ENCODE publicity fiasco.
Here's the forum ...
AskScience Special AMA: We are the Encyclopedia of DNA Elements (ENCODE) Consortium. Last week we published more than 30 papers and a giant collection of data on the function of the human genome. Ask us anything!
It's interesting to see how some of the consortium members are responding to criticism. My personal view is that none of them seem to be very knowledgeable about genome biology and the work that has been published over the past 40 years.
Last week's molecule was N-formylmethionyl-tRNAfMet [Monday's Molecule #188]. Only three people came close. The winner was Mikkel Rasmussen.
Name these molecules. One of them has a well-known common name that you have to include in your answer. The other one also has a common name but you don't have to find it. You have to give the complete formal names of each molecule. Do you know the significance of these two molecules?
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
There are several dozen scientists and graduate students in my department studying various aspects of protein evolution, structure, and folding. They've published dozens of papers but, apparently, they never realized that what they're studying is impossible. Douglas Axe, a leading Intelligent Design Creationist, sets them straight.
[Hat Tip: Evolution News & Views: Axe: Vive la Différence]
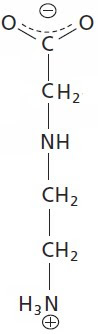
Last week's molecule was the core nucleosome complex [Monday's Molecule #186] and nobody who was eligible for a win got it! That's quite shocking. Here's an easy one for today.
Name this molecule, including the name of the "R" group. You'll have to guess but there's really only one possibility in living cells. Don't forget, I need the full name of the most likely molecule given the partial structure that you see.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
Last week's molecules were the four common nuleosides in DNA [Monday's Molecule #185]. The winner was Matt Talarico.
This week I'm asking you to identify a complex structure made up of eight different components (top) plus one other (bottom). Name the structure making sure to be as specific as possible, Name the none components.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Read more »
Lots of people don't understand the Central Dogma of Molecular Biology and that's probably why there are so many articles announcing its death. The article and book by James Shapiro is just one example [Revisiting the Central Dogma in the 21st Century].
The correct version of the Central Dogma of Molecular Biology is .... [see Basic Concepts: The Central Dogma of Molecular Biology]
... once (sequential) information has passed into protein it cannot get out again (F.H.C. Crick, 1958)
The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred from protein to either protein or nucleic acid. (F.H.C. Crick, 1970)
Eugene Koonin has an article in Biology Direct entitled Does the central dogma still stand (Koonin, 2012).
Read more »
Last week's molecule was warfarin, a rat poison with another role [Monday's Molecule #184]. The winner was Matt McFarlane.
This week we're in the middle of the ENCODE/junk DNA controversy. A dispute that reveals a serious lack of knowledge of fundamental concepts in biochemistry. I'm going to go back to basics today and ask you to name these four molecules. Be careful, I'm going to insist that you use the correct unambiguous names. Name them in order from upper left to upper right to lower left then lower right.
Post your answer as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Some past winners are from distant lands so their chances of taking up my offer of a free lunch are slim. (That's why I can afford to do this!)
In order to win you must post your correct name. Anonymous and pseudoanonymous commenters can't win the free lunch.
Winners will have to contact me by email to arrange a lunch date. Please try and beat the regular winners. Most of them live far away and I'll never get to take them to lunch. This makes me sad.
Comments are now open.
UPDATE: The molecules are deoxyadenosine, deoxyguanosine, deoxycytidine, deoxythymidine. This week's winner is Matt Talarico. Matt should contact me by email.
Winners
Nov. 2009: Jason Oakley, Alex Ling
Oct. 17: Bill Chaney, Roger Fan
Oct. 24: DK
Oct. 31: Joseph C. Somody
Nov. 7: Jason Oakley
Nov. 15: Thomas Ferraro, Vipulan Vigneswaran
Nov. 21: Vipulan Vigneswaran (honorary mention to Raul A. Félix de Sousa)
Nov. 28: Philip Rodger
Dec. 5: 凌嘉誠 (Alex Ling)
Dec. 12: Bill Chaney
Dec. 19: Joseph C. Somody
Jan. 9: Dima Klenchin
Jan. 23: David Schuller
Jan. 30: Peter Monaghan
Feb. 7: Thomas Ferraro, Charles Motraghi
Feb. 13: Joseph C. Somody
March 5: Albi Celaj
March 12: Bill Chaney, Raul A. Félix de Sousa
March 19: no winner
March 26: John Runnels, Raul A. Félix de Sousa
April 2: Sean Ridout
April 9: no winner
April 16: Raul A. Félix de Sousa
April 23: Dima Klenchin, Deena Allan
April 30: Sean Ridout
May 7: Matt McFarlane
May 14: no winner
May 21: no winner
May 29: Mike Hamilton, Dmitri Tchigvintsev
June 4: Bill Chaney, Matt McFarlane
June 18: Raul A. Félix de Sousa
June 25: Raul A. Félix de Sousa
July 2: Raul A. Félix de Sousa
July 16: Sean Ridout, William Grecia
July 23: Raul A. Félix de Sousa
July 30: Bill Chaney and Raul A. Félix de Sousa
Aug. 7: Raul A. Félix de Sousa
Aug. 13: Matt McFarlane
Aug. 20: Stephen Spiro
Aug. 27: Raul A. Félix de Sousa
Sept. 3: Matt McFarlane
Sept. 10: Matt Talarico
Here's an excellent example of what's wrong with the way the ENCODE Consortium is interpreting their data. Congratulations to Michael Eisen! I wish I had said this: A neutral theory of molecular function.1
Read the whole thing very carefully and heed the lesson. Here's a excerpt,
I think a lot about Kimura, the neutral theory, and the salutary effects of clear null models every time I get involved in discussions about the function, or lack thereof, of biochemical events observed in genomics experiments, such as those triggered this week by publications from the ENCODE project.
It is easy to see the parallels between the way people talk about transcribed RNAs, protein-DNA interactions, DNase hypersensitive regions and what not, and the way people talked about sequence changes PK (pre Kimura). While many of the people carrying out RNA-seq, ChIP-seq, CLIP-seq, etc… have been indoctrinated with Kimura at some point in their careers, most seem unable to apply his lesson to their own work. The result is a field suffused with implicit or explicit thinking along the following lines:
I observed A bind to B. A would only have evolved to bind to B if it were doing something useful. Therefore the binding of A to B is “functional”.
One can understand the temptation to think this way. In the textbook view of molecular biology, everything is highly regulated. Genes are transcribed with a purpose. Transcription factors bind to DNA when they are regulating something. Kinases phosphorylate targets to alter their activity or sub-cellular location. And so on. Although there have always been lots of reasons to dismiss this way of thinking, until about a decade ago, this is what the scientific literature looked like. In the day where papers described single genes and single interactions, who would bother to publish a paper about a non-functional interaction they observed?
But experimental genomics blew this world of Mayberry molecular biology wide open. For example, when Mark Biggin and I started to do ChIP-chip experiments in Drosophila embryos, we found that factors were binding not just to their dozen or so non-targets, but the thousands, and in some cases tens of thousands of places across the genome. Having studied my Kimura, I just assumed that the vast majority of these interactions had evolved by chance – a natural, essential, consequence of the neutral fixation of nucleotide changes that happened to create transcription factor binding sites. And so I was shocked that almost everyone I talked to about this data assumed that every one of these binding events was doing something – we just hadn’t figured out what yet.
.....
Rather than assuming – as so many of the ENCODE researchers apparently do – that the millions (or is it billions?) of molecular events they observe are a treasure trove of functional elements waiting to be understood, they should approach each and every one of them with Kimurian skepticism. We should never accept the existence or a molecule or the observation that it interacts with something as prima facia evidence that it is important. Rather we should assume that all such interactions are non-functional until proven otherwise, and develop better, compelling, ways to reject this null hypothesis.
Read the comments, especially the one from former colleague Chris Hogue on how to interpret phosphorylation of proteins and signal transduction. That's not going to be popular in my department!
I just have one small quibble with Michael's post. Not all textbooks describe the cell as if it were a finely tuned Swiss watch and not all textbooks take an adaptationist approach to evolution. Mine doesn't.
1. As a result of this post I've now relegated Jonathan Eisen to "brother of Michael Eisen" rather than the other way around. Sorry, Jonathan.
The Nature issue containing the latest ENCODE Consortium papers also has a New & Views article called "Genomics: ENCODE explained" (Ecker et al., 2012). Some of these scientist comment on junk DNA.
For exampleshere's what Joseph Ecker says,
One of the more remarkable findings described in the consortium's 'entrée' paper is that 80% of the genome contains elements linked to biochemical functions, dispatching the widely held view that the human genome is mostly 'junk DNA'. The authors report that the space between genes is filled with enhancers (regulatory DNA elements), promoters (the sites at which DNA's transcription into RNA is initiated) and numerous previously overlooked regions that encode RNA transcripts that are not translated into proteins but might have regulatory roles.
And here's what Inês Barroso, says,
The vast majority of the human genome does not code for proteins and, until now, did not seem to contain defined gene-regulatory elements. Why evolution would maintain large amounts of 'useless' DNA had remained a mystery, and seemed wasteful. It turns out, however, that there are good reasons to keep this DNA. Results from the ENCODE project show that most of these stretches of DNA harbour regions that bind proteins and RNA molecules, bringing these into positions from which they cooperate with each other to regulate the function and level of expression of protein-coding genes. In addition, it seems that widespread transcription from non-coding DNA potentially acts as a reservoir for the creation of new functional molecules, such as regulatory RNAs.
If this were an undergraduate course I would ask for a show of hands in response to the question, "How many of you thought that there did not seem to be "defined gene-regulatory elements" in noncoding DNA?"
I would also ask, "How many of you have no idea how evolution could retain "useless" DNA in our genome?" Undergraduates who don't understand evolution should not graduate in a biological science program. It's too bad we don't have similar restrictions on senor scientists who write News & Views articles for Nature.
Jonathan Pritchard and Yoav Gilad write,
One of the great challenges in evolutionary biology is to understand how differences in DNA sequence between species determine differences in their phenotypes. Evolutionary change may occur both through changes in protein-coding sequences and through sequence changes that alter gene regulation.
There is growing recognition of the importance of this regulatory evolution, on the basis of numerous specific examples as well as on theoretical grounds. It has been argued that potentially adaptive changes to protein-coding sequences may often be prevented by natural selection because, even if they are beneficial in one cell type or tissue, they may be detrimental elsewhere in the organism. By contrast, because gene-regulatory sequences are frequently associated with temporally and spatially specific gene-expression patterns, changes in these regions may modify the function of only certain cell types at specific times, making it more likely that they will confer an evolutionary advantage.
However, until now there has been little information about which genomic regions have regulatory activity. The ENCODE project has provided a first draft of a 'parts list' of these regulatory elements, in a wide range of cell types, and moves us considerably closer to one of the key goals of genomics: understanding the functional roles (if any) of every position in the human genome.
The problem here is the hype. While it's true that the ENCODE project has produced massive amounts of data on transcription binding sites etc., it's a bit of an exaggeration to say that "until now there has been little information about which genomic regions have regulatory activity." Twenty-five years ago, my lab published some pretty precise information about the parts of the genome regulating activity of a mouse hsp70 gene. There have been thousands of other papers on the the subject of gene regulatory sequences since then. I think we actually have a pretty good understanding of gene regulation in eukaryotes. It's a model that seems to work well for most genes.
The real challenge from the ENCODE Consortium is that they question that understanding. They are proposing that huge amounts of the genome are devoted to fine-tuning the expression of most genes in a vast network of binding sites and small RNAs. That's not the picture we have developed over the past four decades. If true, it would not only mean that a lot less DNA is junk but it would also mean that the regulation of gene expression is fundamentally different than it is in E. coli.
[Image Credit: ScienceDaily: In Massive Genome Analysis ENCODE Data Suggests 'Gene' Redefinition.
Ecker, J.R., Bickmore, W.A., Barroso, I., Pritchard, J.K. (2012) Genomics: ENCODE explained. Nature 489:52-55. [doi:10.1038/489052a]
Yoav Gilad
& Eran Segal
Ed Yong is a science journalist and usually he's a very good one. This time, however, he should have gotten the other side of the story.
Ed interviewed Ewan Birney for a story on the function of sequences in the human genome [ENCODE: the rough guide to the human genome].
According to ENCODE’s analysis, 80 percent of the genome has a “biochemical function”. More on exactly what this means later, but the key point is: It’s not “junk”. Scientists have long recognised that some non-coding DNA probably has a function, and many solid examples have recently come to light. But, many maintained that much of these sequences were, indeed, junk. ENCODE says otherwise. “Almost every nucleotide is associated with a function of some sort or another, and we now know where they are, what binds to them, what their associations are, and more,” says Tom Gingeras, one of the study’s many senior scientists.
And what’s in the remaining 20 percent? Possibly not junk either, according to Ewan Birney, the project’s Lead Analysis Coordinator and self-described “cat-herder-in-chief”. He explains that ENCODE only (!) looked at 147 types of cells, and the human body has a few thousand. A given part of the genome might control a gene in one cell type, but not others. If every cell is included, functions may emerge for the phantom proportion. “It’s likely that 80 percent will go to 100 percent,” says Birney. “We don’t really have any large chunks of redundant DNA. This metaphor of junk isn’t that useful.”
The creationists are going to love this.
You blew it Ed Yong. Why didn't you ask him about the 50% of our genome containing DEFECTIVE transposons and the 2% that's pseudogenes, just for starters? Then you could ask him why he believes that all intron sequences (about 20% of our genome) are functional [What's in Your Genome?].
"Almost every nucleotide ..."? Gimme a break. Don't these guys read the scientific literature?
This is going to make my life very complicated.
Last week's molecule was raltitrexid, an anti-cancer drug [Monday's Molecule #183]. The winner was Raul A. Félix de Sousa. Raul has won ten times since I restarted Monday's Molecule last November.
This week's molecule is another strange-looking molecule with a very specific purpose. Identify the molecule and its role in mammals.
Post your answers as a comment. I'll hold off releasing any comments for 24 hours. The first one with the correct answer wins. I will only post mostly correct answers to avoid embarrassment. The winner will be treated to a free lunch.
There could be two winners. If the first correct answer isn't from an undergraduate student then I'll select a second winner from those undergraduates who post the correct answer. You will need to identify yourself as an undergraduate in order to win. (Put "undergraduate" at the bottom of your comment.)
Some past winners are from distant lands so their chances of taking up my offer of a free lunch are slim. (That's why I can afford to do this!)
In order to win you must post your correct name. Anonymous and pseudoanonymous commenters can't win the free lunch.
Winners will have to contact me by email to arrange a lunch date. Please try and beat the regular winners. Most of them live far away and I'll never get to take them to lunch. This makes me sad.
Comments are invisible for 24 hours. Comments are now open.
UPDATE: The molecule is warfarin or Coumadin®, a rat poison and an anticoagulant. It's a competitive inhibitor of vitamin K reductase and this blocks blood clotting. The winner is Matt McFarlane, one of the few people who can actually collect a free lunch. Please contact me by email.
Winners
Nov. 2009: Jason Oakley, Alex Ling
Oct. 17: Bill Chaney, Roger Fan
Oct. 24: DK
Oct. 31: Joseph C. Somody
Nov. 7: Jason Oakley
Nov. 15: Thomas Ferraro, Vipulan Vigneswaran
Nov. 21: Vipulan Vigneswaran (honorary mention to Raul A. Félix de Sousa)
Nov. 28: Philip Rodger
Dec. 5: 凌嘉誠 (Alex Ling)
Dec. 12: Bill Chaney
Dec. 19: Joseph C. Somody
Jan. 9: Dima Klenchin
Jan. 23: David Schuller
Jan. 30: Peter Monaghan
Feb. 7: Thomas Ferraro, Charles Motraghi
Feb. 13: Joseph C. Somody
March 5: Albi Celaj
March 12: Bill Chaney, Raul A. Félix de Sousa
March 19: no winner
March 26: John Runnels, Raul A. Félix de Sousa
April 2: Sean Ridout
April 9: no winner
April 16: Raul A. Félix de Sousa
April 23: Dima Klenchin, Deena Allan
April 30: Sean Ridout
May 7: Matt McFarlane
May 14: no winner
May 21: no winner
May 29: Mike Hamilton, Dmitri Tchigvintsev
June 4: Bill Chaney, Matt McFarlane
June 18: Raul A. Félix de Sousa
June 25: Raul A. Félix de Sousa
July 2: Raul A. Félix de Sousa
July 16: Sean Ridout, William Grecia
July 23: Raul A. Félix de Sousa
July 30: Bill Chaney and Raul A. Félix de Sousa
Aug. 7: Raul A. Félix de Sousa
Aug. 13: Matt McFarlane
Aug. 20: Stephen Spiro
Aug. 27: Raul A. Félix de Sousa
Sept. 3: Matt McFarlane
Tim Radford reviews The Selfish Gene by Richard Dawkins [The Selfish Gene by Richard Dawkins – book review]. The review is a bit late—the book was published in 1976—but I suppose the old adage of "better late than never" applies.
Actually it's not as bizarre as you might think. Lot's of people don't understand the ideas that Dawkins was pushing. He was mostly pointing out that evolution is a phenomenon that takes places at the level of genes and populations. Dawkins tweets that Rafford "gets it" in his review.
Lovely retrospective review of The Selfish Gene by Tim Radford, the Guardian's distinguished science writer. He gets it.
Read more »
nature science for kids,nature science definition,nature science articles,nature science jobs,nature science museum,nature science projects,nature science magazine,nature science journal
nature science for kids,nature science definition,nature science articles,nature science jobs,nature science museum,nature science projects,nature science magazine,nature science journal